Six years ago, UCLA biostatistician Steve Horvath realized the potential value of an aging clock and set out to measure human age using methylation markers in DNA from across the body. He used statistical pattern-recognition software to look for relationships between a person’s age and the methylation state of his DNA. Methylation is the best-studied of the epigenetic markers that control which genes are turned on and off, and different sets of genes are active at different stages of life.

thanks to the Horvath lab for this image
Age is an important predictor because the diseases that kill most of us all occur in a highly age-dependent way. In fact, the risks for cancer, heart disease, and Alzheimer’s disease all rise exponentially with age.
One statistical result from the original Horvath clock has a profound implication which aging researchers have been slow to take to heart: The Horvath clock was derived with statistical methods that looked only at chronological age. The algorithm was optimized to produce the best estimate of a person’s calendar age. Of course, age by the calendar is a good predictor of a person’s risk of death. In Americans over 40, the probability of death doubles every 8 years.
We should expect that since the Horvath clock is well-correlated to age and age is well-correlated to mortality, the Horvath clock should be correlated to mortality. (This isn’t guaranteed mathematically except when the two separate correlations are strong.) The interesting twist is this: The Horvath clock is more tightly correlated with mortality than age itself. The clock algorithm was derived from chronological age, so the math knows only about calendar years. But the clock algorithm predicts mortality better than age itself.
We can conclude that this extra accuracy of the methylation clock derives not from math but from biology. The message is that methylation is linked to the biological process of aging. Methylation changes don’t just happen over time; they are coupled to whatever it is that causes the risk of death to rise, linked, in other words, to aging itself.
With more recent developments in the clock, this conclusion gets stronger, and also stranger.
2017 The Zhang Clock
Yan Zhang of the German National Cancer Inst in Heidelberg has developed a methylation-based computation of mortality risk which is based on historic samples of blood from 406 people who died over a 15-year period and from 1,000 demographically-matched control.
They identify 58 sites that were tightly coupled to mortality. In 49 out of 58, less methylation was associated with a higher risk of death, and in the other 9, more methylation led to higher risk of death. (More methylation corresponds to less gene expression. The message is that increase in age-related mortality is due more to turning on genes that destroy us than to silencing genes that protect us.)
None (count ’em–zero!) of the 58 were incorporated in any of the previously published aging clocks (by Horvath and Hannum). What do we make of this? Age is associated with mortality more closely than any other biological indicator, and in fact mortality risk rises exponentially with age. And yet Zhang et al set out to look for methylation sites most closely associated with mortality risk, Horvath et al set out to look for methylation sites most closely associated with chronological age, and there was zero overlap between the sites they identified! In fact, less than half the sites they identified (23/58) had statistically significant correlations with age at all.
The recently established epigenetic clock (DNAm age) has received growing attention as an increasing number of studies have uncovered it to be a proxy of biological ageing and thus potentially providing a measure for assessing health and mortality. Intriguingly, we targeted mortality-related DNAm changes and did not find any overlap with previously established CpGs that are used to determine the DNAm age.
Part of the explanation may be that Zhang’s study was conducted in an older population (median age=62) at higher risk of death, and that the Horvath clock to which he compared it was designed to generally reflect age, from womb to tomb. Zhang says, “Methylation levels were measured on average 8.2 years before dying.”
Zhang’s mortality risk estimator is a count of how many of the 10 most telling methylation sites are in the “worst” quartile of his test population. (The “worst” quartile is the highest quartile for some and the lowest quartile for other sites.) A score of 5 corresponds to a 7-fold increase in mortality risk. This qualifies the Zhang score as one of the most powerful risk indicators that we have (don’t tell Aetna). For comparison, a BMI of 35 qualifies as “obese” and corresponds to a mortality risk ratio of only 1.36. Hemoglobin A1C, and HDL are common indicators of health status in older adults, and all of these have marginal associations with age-adjusted mortality. C-reactive protein and IL6 are blood markers of inflammation, and they were associated with risk ratios of 1.6 and 1.9, respectively. By this standard, the Zhang score is a big step forward.
Methylation is presumed to be under the body’s programmatic control. There are two reasons that methylation might be powerfully associated with mortality. First, some changes in methylation may be an indication of an acute response to some life-threatening stress; second, some changes in methylation may be part of an intrinsic death program associated with age. My guess is that there is some of each going on, but probably more of the former, since (as I said) only 23 of the 58 sites are significantly correlated with age.
Another curious fact: the methylation sites associated with smoking provided a better indicator mortality risk than was smoking itself. More about this below.
2018: The Levine Clock
Morgan Levine, working with Horvath at UCLA, developed a second-generation clock last year based on mortality and morbidity data as well as chronological age. The Levine clock was optimized with hindsight, factoring in age-related disease that occurred years after the blood was sampled.
Levine and her team worked in two stages. First, they developed a measure they call “phenotypic age” which includes age itself plus 9 modifiers that contribute to mortality risk.
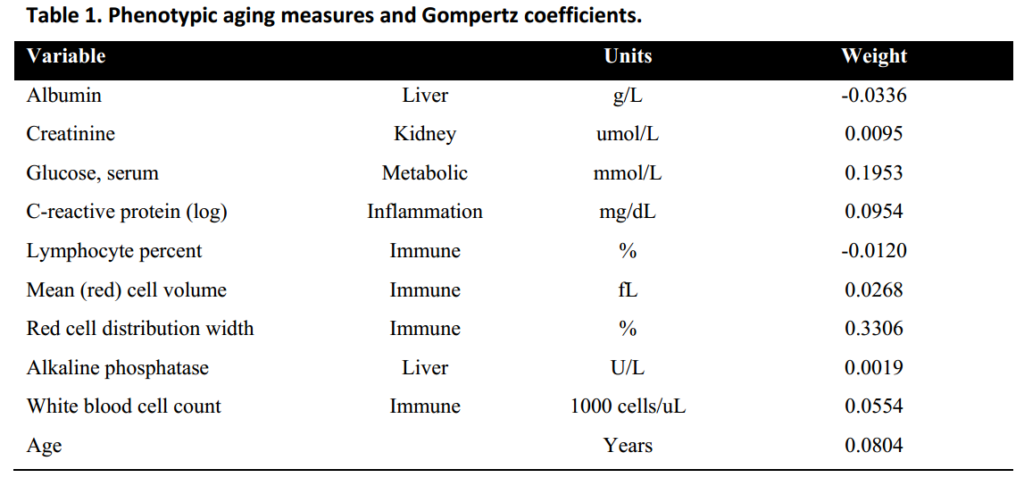
Albumin: dissolved proteins in the plasma, including hormones and other signal molecules.
Creatinine: this is a waste product cleared by the kidneys, thus a high value suggests kidney malfunction; but it can be confounded by exercise, which raises creatinine.
Glucose: blood sugar rises with Type 2 diabetes and loss of insulin sensitivity.
C-reactive protein: this is a measure of systemic inflammation.
Lymphocyte %: the most common types of white blood cells.
Mean red cell volume (MCV): the average size of red blood cells
Red cell distribution width (RDW): standard deviation of the above
Alkaline phosphatase (ALP): this is elevated in liver disease, including cancers and hepatitis.
White blood cell count: total white blood cells of all types
The list surprised me. This was not a popularity contest; it was developed from statistical association with mortality, with no prejudices up front. I was not surprised to see glucose and CRP in the list (though I would have thought they would substitute A1C for glucose, because A1C is more stable, while glucose varies from hour to hour). I would have thought to find HDL and IL-6 in the list, and I was particularly surprised to see the strongest weighting was Red cell distribution width, which I had not heard of. RDW is measured as the standard deviation in volume of individual red blood cells (erythrocytes). It turns out that small red blood cells are a symptom of diabetes, while high RDW scores are associated with cancer and heart disease. There’s a modest association between RDW and Alzheimer’s Dementia.
Also curious: total white blood cell count is positively associated with aging diseases, while lymphocytes, a subset of white blood cells, has a negative association. So, what are the white blood cells that are not lymphocytes? These comprise neutrophils, eosinophils, monocytes, and basophils. Large quantities of these are a warning of bad health to come. Neutrophils are the largest category among these, and they are part of the body’s innate defense against cancer and infections. Lymphocytes, on the other hand, comprise natural killer (NK) cells and T- and B-cells. NK cells are part of the innate immune system, while T-and B-cells are part of the adaptive immune system, but all of these are indicative of good health and long life.
All these components were put together by Levine et al to form their measure of phenotypic age. The team then went on to stage two, looking for methylation sites that correlate best with their newly-defined measure of phenotypic age. 513 sites were incorporated in their computation (see below). This can be confusing: PhenoAge is the measure derived from the above 9 blood tests + chronological age. DNAm PhenoAge is the methylation clock derived from the PhenoAge blood test.
The resulting PhenoAge methylation clock (DNAm PhenoAge) correlates only about 75% with chronological age (compared to 94% for the original Horvath clock). But DNAm PhenoAge predicts mortality and morbidity far better than either chronological age or the original Horvath clock. As you might expect, the methylation clock which was derived from the newly-invented PhenoAge measure does not predict mortality rates as well as PhenoAge itself, from which it was derived. This is expected because the DNAm PhenoAge clock is targeted directly to predict PhenoAge, and only indirectly to predict mortality. I am only making a point of this because the story is different and surprising in the case of the new GrimAge methylation clock–described next week.
Fifty sites vs Five Hundred
The first step in producing a clock is to produce a list of individual methylation sites in order of how tightly they correlate with age. If you construct a clock out of the first few, you get the best correlation and the most accurate measure of age. But the measure is fragile, and the accuracy may be illusory. When selecting a few items out of a list of hundreds of thousands, there will usually be accidents and outliers, statistical flukes. By including more sites assures that the overall age measure is not unduly affected by any one site, so if a few of the correlations turn out to be statistical errors, the overall average is still quite good. Horvath has generally chosen to be conservative and sacrifice some accuracy for robustness.
Next week, the new GrimAge clock…
Methylation measurements have provided the most accurate measure of age and prediction of age-related disease, head and shoulders above other measures. But can we do even better if we supplement methylation data with other things we know about a person–not just other blood tests, but life style factors. When I visited Horvath last summer, he introduced me to his post-doc Ake Lu, who was working on a composite clock, based on this thinking: methylation plus. That was the origin of the GrimAge clock.
Josh Mitteldorf


